Glues! Glues? Everywhere?
2nd February 2024
There is increasing excitement, and indeed success, around the search for molecular glues which induce target degradation, or potentially other, interesting downstream biology by persuading two proteins who don’t normally hang around together to do just that. This should perhaps not be surprising as nature uses molecular glues to regulate various biological systems (auxin, cyclosporine, FK506 etc) so it’s likely that HTS collections already contain many of these glues just waiting to be discovered.
When thinking of the high protein load in a typical cell (up to 300mg/ml), it’s perhaps more surprising that proteins don’t go round interacting with each other in non-specific ways in the absence of glues given the sometimes partially hydrophobic nature of some protein surface sites swimming in a largely aqueous medium. Clearly, millennia of evolution have done their best to minimise these lawless interactions in favour of the well-controlled associations which underpin many biological processes. However, many proteins must be on the cusp of wanting to interact with other things so by binding a small molecule, they can be persuaded to do this via new interacting surfaces. Proteins can tip into lots of interactions with other proteins without small molecule help of course – just look at all the amyloid and other aggregation-prone proteins which underpin many disease-causing processes.
Rationally designing small molecule glues which persuade two proteins to get cosy with each other is not trivial though many groups are now grappling with this challenge. The recent report from the Novartis team (BioRXiv preprint here) however shows that glues can also be found in more familiar places.
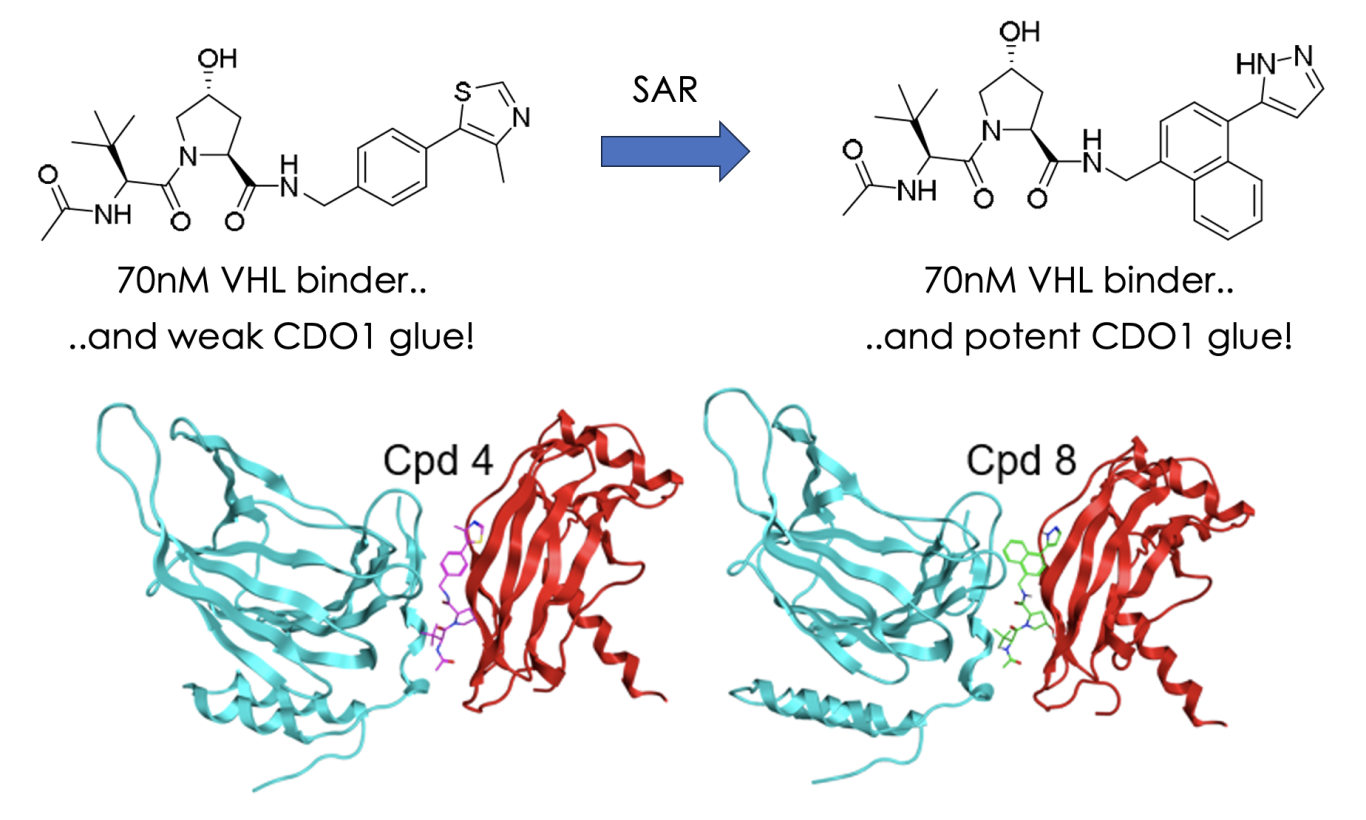
Starting from the very well known VHL-binding hydroxyproline ligands, they asked the question what other proteins can the VHL-ligand complex recruit? Perhaps surprisingly, the “standard” VHL ligand (above, left – compound 4 in their manuscript) when in complex with VHL, but not alone, recruited the protein cysteine dioxygenase 1 (CDO1) leading to its proteasomal degradation with a respectable DC50 of 350nM via a co-operative ternary complex. Some quick SAR improved this DC50 to 8nM (above, right – compound 8).
Although by the authors own admission degradation of CDO1 has no obvious therapeutic application, impressively, it was the only protein to be degraded from >9500 assessed in expression proteomics showing the specificity of the glue. The authors also include a range of mechanistic studies to confirm that the VHL binding analogs are bona fide CDO1 glues, culminating in ternary complex x-ray crystal structures (shown above) nicely showing the binding at the interface and highlighting key interactions. Using this information, they identified the bipartite degron motif on the surface of CDO1 which allows the glue interaction and searched for this in other proteins to find other “glue-able” targets but were unsuccessful.
Does this mean that every glue-able interface is unique or rather that the motifs can only be described using a more complex combination of primary sequence segments overlayed with three-dimensional topology of a suitable conformation in a dynamic protein surface? This is where the predictive algorithms can really earn their worth if the compute is possible. Cryptically the authors allude to identifying VHL-based glues of other, unnamed targets though it wasn’t clear if this was through rational design or analog screening – we’ll have to be patient for more of those stories to appear.
This lovely work highlights a few interesting and potentially important observations.
Glues are probably more common than you might think: All of the analogs 4-8 in the paper showed some evidence of VHL-glue-CDO1 ternary complex formation via one or more proximity assays.
However, only a subset of these resulted in the desired function, in this case degradation. From the limited dataset, it appears some threshold of ternary complex stability (described by eg Amax) needs to be reached before they become functionally productive. The non-degradative ternary complexes may however have facilitated other biology of course though the authors didn’t characterise this.
The data also nicely show that very small changes can have big impacts on the ability of molecules to be functional glues. Adding just a 4 carbon atom degron motif (phenyl to naphthyl) to compound 4 to give compound 8 gave >50 fold increase in glue activity. Contrarily, removing a single carbon from compound 4 (to give compound 3) gave a weaker ternary complex and abolished all degradation. This high sensitivity effect is often seen in glue space: Adding 4 atoms to the ubiquitous BRD4 inhibitor JQ1 yields DCAF16-based glue degrader GNE-0011 while adding a single 2-pyridyl unit to CDK12 inhibitor roscovitine gave CR8, a DDB1-recruiting molecular glue collateral cyclin K degrader. The extensive games which can be played with lenalidomide-based CRBN binders to make a plethora of glues to different targets is of course well described.
It's likely that a range of known inhibitors already act as molecular glue to recruit an effector protein – if this effector is related to the ubiquitin proteasome system, the effect may manifest as target degradation though if the effector is from a different protein family, the functional consequence could be entirely different and open up a wide range of hitherto inaccessible pharmacology.
And, even if current inhibitors possess even only weak glue-like character, making small changes to them to enable display of a hydrophobic patch may be all that is needed to stabilise ternary complexes and lead to useful function.
There’s a lot to discover in glue-like space – some of it will be rational, some of it may be serendipitous or poorly understood (the IMiD drugs were shown to be clinically active long before their full glue MoA was elucidated of course) but all of it will be interesting. We may not be interested in degrading CDO1 at this time, but by learning lessons, we’re more likely to find agents that mediate pharmacology that we’ve never been able to access before and that must be good for the development of new therapeutics.
We need your consent to load the translations
We use a third-party service to translate the website content that may collect data about your activity. Please review the details in the privacy policy and accept the service to view the translations.

